Anna M. Pyle
Professor of Chemistry
Howard Hughes Medical Institute Investigator
Member of Yale faculty since 2002
Research Interests
Research
Anna Marie Pyle studies RNA structure and RNA recognition by protein enzymes. Her lab uses a combination of experimental biochemistry, crystallography and computation to study the architectural features of large RNA molecules, such as self-splicing introns and other noncoding RNAs. This is accompanied by complementary work on RNA-dependent ATPase enzymes that bind and remodel RNA structures, with an emphasis on proteins that are involved in viral replication and host innate immune response.
In the Pyle lab, we focus on two related questions: (1) How do large RNAs assemble into specific, stable tertiary structures? (2) How is RNA recognized and remodeled by ATP-dependent enzymes in the cell? Our studies involve a combination of solution biochemistry, enzymology, crystallography, and cell-based functional approaches. In parallel, we develop new computational methods for solving and analyzing RNA structures.
Group II Introns and Other Large RNA Tertiary Structures
Our studies of RNA tertiary architecture have focused on group II introns, which are large self-splicing ribozymes that are essential for gene expression in many organisms. Second only to the ribosome in size, group II introns have provided key insights into our understanding of RNA structure and evolution.
Initially, my laboratory used solution biochemistry and enzymology to characterize the chemical reaction mechanisms and architecture of group II introns. While this work yielded important insights into RNA splicing, we required high-resolution information on group II intron structures to define their functions precisely. We therefore spent many years attempting to identity a stable, homogeneous group II intron suitable for structural studies and were finally successful in crystallizing and solving the structure of a group IIC intron from the bacterium Oceanobacillus iheyensis (Oi IIC, ~400 nucleotides in size). This molecule, which is among the largest free RNA structures ever solved, revealed new architectural motifs and novel strategies for catalysis by RNA molecules (Figure 1, left).
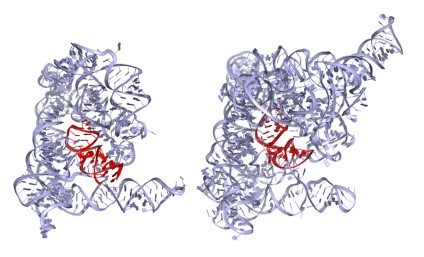 Figure 1. Crystal structure of the Oi group IIC intron (left, 4FAR)
Figure 1. Crystal structure of the Oi group IIC intron (left, 4FAR)
We have since solved the Oi IIC structure as it moves through the stages of splicing, showing that both steps are catalyzed by a conserved RNA stem loop (domain 5, red in Figure 1) that contains a reactive metal ion cluster composed of magnesium and potassium ions. In addition, we recently captured a conformational change that occurs between the two steps of splicing, allowing the active site to exchange splice sites and carry out multistep reactions. Using these structures, we have adapted homology-modeling programs and applied them to RNA, thereby modeling the structures of much larger group II introns, such as the ai5? group IIB intron from yeast mitochondria (Figure 1, right).
Group II introns are particularly useful model systems for understanding the eukaryotic spliceosome, which processes pre-mRNA molecules in the nucleus. It had long been hypothesized that U6 snRNA (small nuclear RNA) within eukaryotic spliceosomes behaves in a manner similar to domain 5 of group II introns. Using our crystal structures as a guide, we created a road map for identifying U6 catalytic groups and we predicted the molecular organization of the splicesomal active site. Recent work at the University of Chicago by Joseph Piccirilli and Jonathan Staley, our colleagues in the spliceosome field, has confirmed our predictions and shown that the spliceosome is a ribozyme that is organized much like a group II intron. This work provides a strong foundation for exploiting the potential of group II introns in gene therapy and for developing group II introns and spliceosomes as therapeutic targets. It also provides the methodologies and strategies needed for solving the structures of even larger RNA molecules, such as long intergenic noncoding RNAs (lincRNAs), that play a central role in epigenetic control and other processes.
Protein Machines for RNA Remodeling and Sensing
Eukaryotic cells express a large family of RNA-dependent ATPases (SF2 ATPases/helicases) that contribute to every aspect of RNA metabolism. Many of these proteins unwind RNA structures during the remodeling of RNA-protein complexes (acting as helicases), while others stabilize RNA structures (behaving as annealing enzymes), and yet others serve as biosensors and signals for the detection of pathogenic RNA (signaling enzymes). These proteins share certain architectural elements, including a common set of conserved domains that selectively bind RNA targets and create an active-site cleft for ATP binding and hydrolysis. The ATP-dependent motions of this cleft are coupled to mechanical funcitons, such as the unwinding of RNA, or domain motions that promote cell signaling. In studying the nanomechanical behavior of these proteins, we have explored new areas of molecular virology and immunology.
We are particularly interested in SF2 RNA helicase enzymes that play a role in the life cycle of viruses. For example, the NS3 helicase from hepatitis C virus (HCV) plays a key role in the replication and packaging of HCV. We have elucidated the stepwise mechanism by which NS3 unwinds RNA molecules, and we have used it as a paradigm for understanding ATP-powered translocation within the SF2 family. We have begun to dissect the network of interactions between NS3 and other components of the HCV replication complex, and we have shown that this multifunctional enzyme plays many roles in HCV pathogenicity.
During early studies of a protein involved in cancer reversion (MDA-5), we identified a subfamily of SF2 proteins that displays highly unusual behavior. The ATPase activity of these proteins, which include proteins MDA-5, RIG-I, and metazoan Dicer, is specifically stimulated by duplex RNA, rather than single-stranded RNA and it is not accompanied by RNA unwinding. Family members such as RIG-I and MDA-5 play a central role in the human innate immune system, and Dicer proteins are key components of small interfering RNA (siRNA)- and microRNA (miRNA)-processing systems. Despite the biological significance of all these proteins, there was no high-resolution information on their structures or RNA binding interfaces and limited information on their enzymology.
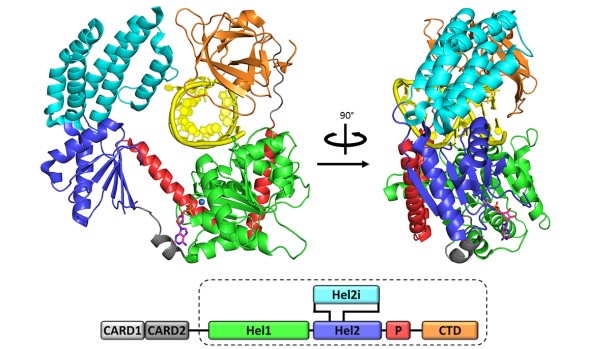 Figure 2: Crystal Structure of RIG-I in complex with RNA hairpin and Adenosine Disphosphate
Figure 2: Crystal Structure of RIG-I in complex with RNA hairpin and Adenosine Disphosphate
We set out to change this with an intensive study of RIG-I, a surveillance protein that detects and responds to viral RNA infection within vertebrate cells. Through in vitro and in vivo experiments, we demonstrated that a 5’-triphosphorylated 10-base pair RNA duplex is sufficient for activating RIG-I and inducing a robust interferon response in vertebrates. We solved the crystal structure of RIG-I in complex with a variety of ligands, revealing an intricate machine that mechanically couples viral RNA binding with ATPase activity and signaling (Figure 2). This work paves the way for the design of new therapeutics that modulate the innate immune response and for new vaccine adjuvants. It also lays the groundwork for mechanistic understanding and pharmacological control of Dicer and related proteins.
Education
B.A. Princeton University, 1985
Ph.D. Columbia University, 1990
Jane Coffin Childs Postdoctoral Fellow, University of Colorado, 1992
Honors
Sterling Professor of Molecular, Cellular and Developmental Biology, Yale University, 2018
Novartis Lecture, University of Pennsylvania, 2018
Gomberg Lecture, University of Michigan, 2018
Jerry A. Weisbach Memorial Lecture, Rockefeller University, 2018
Blavatnik Fund for Innovation Award, New Haven, 2017, 2018, and 2019
Plenary Lecturer, Annual ASMB Meeting, San Diego, CA, 2016
Elected Member, Connecticut Association of Science and Engineering, 2016
Keynote Speaker, Keystone Meeting on Positive-Strand Viruses, 2016
Green Lectureship in Enzymology, University of Wisconsin, Madison, 2016
Distinguished Lecturer, Frontiers in Chemistry, Case Western Reserve University, 2013
Michael Gait Award and Lectureship, Cambridge University, 2013
Distinguished Visitor, European Molecular Biology Laboratory, Heidelberg, 2011
Karl Friedrich Bonhoeffer Lecturer in Biophysics, Göttingen, 2011
Fellow of the American Association for the Advancement of Science, 2007
Endowed Chair, The William Edward Gilbert Professorship, Yale University, 2005
Member of the American Academy of Arts and Sciences, 2005
Mayor’s Award for Excellence in Science and Technology, New York, 2002
Investigator, Howard Hughes Medical Institute, 1997-present
NSF National Young Investigator Award, 1994-1999
Beckman Young Investigator, 1994-1996
Searle Scholars Award, 1993-1996
Irma T. Hirschl and Monique Weill-Caulier Career Scientist Award, 1992-1997
Jane Coffin Childs Memorial Fund Postdoctoral Fellowship, 1990-1992
Pegram Award, Columbia University, 1989
J. Malcolm Miller Teaching Award, Columbia University, 1986
Magna cum laude in Chemistry, Princeton University, 1985
Woodrow Wilson School Program in Science Policy, Princeton University, 1983-1985
Representative Publications
Melissa M. Linehan, Thayne H. Dickey, Emanuela S. Molinari, Megan E. Fitzgerald, Olga Potapova, Akiko Iwasaki and Anna Marie Pyle. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. Feb 21, 2018, 4, E1701854.
Chen Zhao and Anna Marie Pyle. Structural insights into the mechanism of group II intron splicing. Trends Biochem Sci., 2017, 42, 470-482. PMCID: PMC5492998.
Fei Liu, Srinivas Somarowthu and Anna Marie Pyle. Visualizing the secondary and tertiary architectural domains of IncRNA RepA. Nature Chemical Biology, 2017, 13, 282-289. PMID: 28068310.
Nathan Pirakitikulr, Andrew Kohlway, Brett D. Lindenbach and Anna Marie Pyle. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Molecular Cell, 2016, 62, 111-120. PMCID: PMC4826301.
Chen Zhao, K. R. Rajashankar, Marco Marcia and Anna Marie Pyle. Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nature Chemical Biology, 2015, 11, 967-972. PMCID: PMC4651773.
Complete List of Published Work in MyBibliography
Selected Non-Technical Videos:
Science on Saturday: Molecules that Walk, Shuffle, and Carry Things Around
The iBiology Video Series:
RNA Structure
Inside an RNA Splicing Machine
RNA Helicases and RNA-triggered Signaling Machines